Calculating Gene Expression From Exon Arrays
How Does AltAnalyze Determine Gene Expression Levels from Exon Arrays?
While exon arrays probe multiple genomic features (exons, introns, UTRs) often, it is necessary to differentiate transcriptional activity of the gene (gene expression) from alternative modes of transcript expression. Given that many genomic features in or around a gene may not be informative to assess the transcriptional activity of a gene, AltAnalyze employs two strategies to assess gene expression:
- Average the expression of only constitutive probesets (using known transcript annotations - see below)
- Average the expression of all core probesets for a gene
The user can select which method they want AltAnalyze to use, however, in some cases both methods are needed. The first strategy attempts to select only probesets that lack evidence of alternative expression while the second relies on the average of all core probesets, assuming outlier effects will not dominate. In addition to these two strategies, probesets are excluded if they fail to meet certain user defined probeset RMA intensity and detection above background (DABG) threshold (see below). AltAnalyze separately reports gene expression values (ExpressionOutput file) for all genes and predictions of alternative exon expression (AlternativeOutput file). For both of these two files, the analysis requires similar but slightly different methods. These differences are necessary to ensure that for the alternative exon analysis poorly expressed probesets do not contribute to faulty alternative exon predictions and for the gene expression only analysis that all genes have a reported expression value, not just those constitutive expressed probesets. Thus, there are additional constraints in the alternative exon analysis that restrict when gene expression values are reported and from which probesets for very specific biological comparisons.
AltAnalyze Constitutive Probesets
Constitutive exons are those exons which are common to all or nearly all mRNA transcripts for a given gene. This is illustrated in Figure 1, where the black filled boxes are exons or exon regions common to all mRNA transcripts for the same gene.
Figure 1
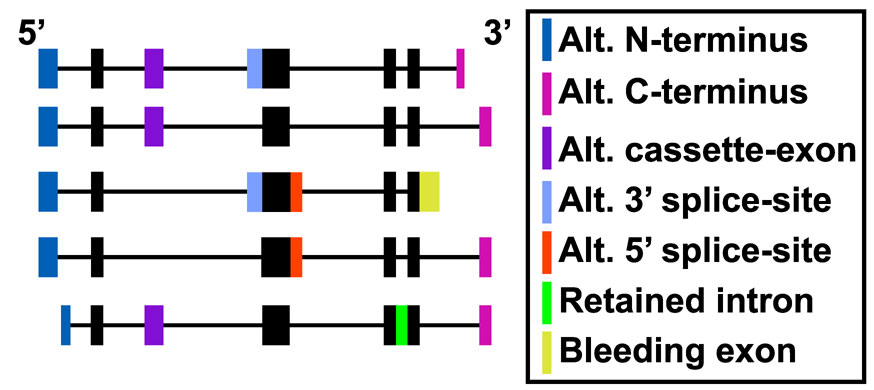
Constitutive exons for a set of mRNA transcripts.
Thus, ideally, a constitutive exon should be the best indicator of gene transcription as opposed to alternative splicing, alternative promoter activity or alternative cleavage and polyadenylation. Affymetrix probesets hybridize to distinct exons and exon regions, thus it is possible to determine which probesets occur in these predicted constitutive transcript regions. AltAnalyze constitutive probeset annotations are assigned when the database is built, thus, these annotations do not change from analysis to analysis. This is in contrast to other methods, such as MADS[Xing2008] where constitutive probesets are chosen based on which probesets have the most stable expression across a number of experiments. The most up-to-date version of AltAnalyze (version 1.14) determine constitutive exons by comparing the composition of exon regions delineated from all Ensembl and UCSC database mRNAs among those transcripts to find those exon regions present among the most transcripts. In previous versions of AltAnalyze, constitutive probesets were selected by using annotations from the Affymetrix probeset.csv annotation file for the respective exon array (e.g., HuEx-1_0-st-v2.na23.hg18.probeset.csv), by identifying which probesets are most frequent to all mRNAs.
Currently, constitutive exon regions are defined by counting the number of unique Ensembl and UCSC mRNA transcripts (based on structure) associated with each unique exon region. If multiple exon regions have the same number of transcript associations, then these are grouped. The grouped exon regions that contained the most transcripts for the gene are defined as constitutive exon regions. If only one exon region is considered constitutive then the grouped exon regions with the second highest ranking (based on number of associated transcripts) are included as constitutive (when there at least 3 ranks). Any probeset that overlaps with a constitutive exon region is assigned the annotation "constitutive" and is used for determining gene expression, when that probeset is deemed expressed and when the user chooses "constitutive probesets" as the mode for gene expression calculation.
Calculating Gene Expression for the ExpressionOutput File
Gene expression values for all genes are saved to the folder ExpressionOutput for the file with the prefix "DATASET". This file can have any number of arrays and biological groups, however, comparison statistics between user defined pairwise groups are reported. In this file, gene expression values are reported for each array and array group for each Ensembl gene. These are the averaged log2 RMA intensities from one or more probesets deemed useful for determining gene expression. The user can choose constitutive or core probesets for "Determine gene expression levels using", under the "Expression Analysis Parameters" menu. Core is all AltAnalyze defined core probesets (which are Affymetrix core plus those found in Ensembl and UCSC mRNAs).
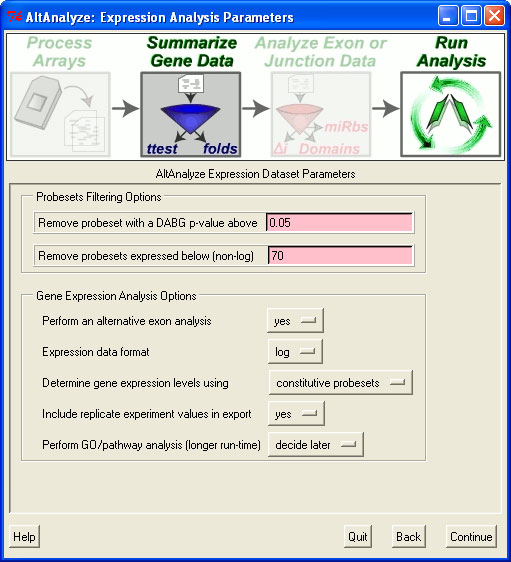
If the default no is selected, AltAnalyze will use only constitutive annotated probesets when possible. If yes...
Additional Information
For more details, see Constitutive and Gene Expression Calculation.