Pathway Visualization in AltAnalyze
Pathway Analyses in AltAnalyze refer to:
- Enrichment of biological categories (e.g., Ontologies, pathways, transcrption factor targets, microRNA binding sites, cell-type markers, custom sets).
- Visualization of WikiPathways with regulated genes or metabolites.
Users can obtain visualized WikiPathways using either the Pathway Analysis menu (found in Additional Analyses) or from the Pathway Visualization menu option (also in Additional Analyses).
AltAnalyze will visualize WikiPathways using either pre-computed results or user provided datasets. In addition to gene IDs, protein IDs and metabolites can be visualized on these pathway maps. Input files are in the GO-Elite format which is automatically saved when you run a workflow analysis (GO-Elite/regulated folder).
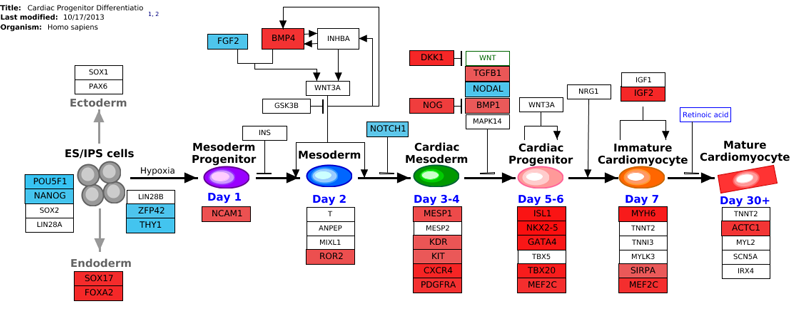
Preparing Your ID List
If you are not using pre-built criterion lists from AltAnalyze (see the folder GO-Elite/regulated), you must create these files. This file should be created as a tab-delimited text file with the IDs of interest (column 1), the GO-Elite system-code (column 2) and an optional log2 fold change (column 3). If not fold change data is present, the nodes in the pathway will be colored yellow, otherwise, positive values will be colored red and the negative blue as shown above. The system code is a two letter code which indicates the ID system being applied. These codes are listed here. AltAnalyze can try to guess the system code if not present but this often does not work. Once finished, your file should look like this example.
| Source Identifier (REQUIRED) | SystemCode (RECOMMENDED) | Fold (OPTIONAL) |
|---|---|---|
| j05479_s_at | X | 3.2 |
| Msa.33069.0_s_at | X | -2.1 |
Pathway Visualization Menu
From the AltAnalyze main menu, select your species and platform. Instead of indicating the platform you can also select "Other ID" as the data type and the gene system you are analyzing as the "platform ID". Select Continue and then Additional Analyses. From here, select Pathway Visualization to see the options for loading your input ID list.
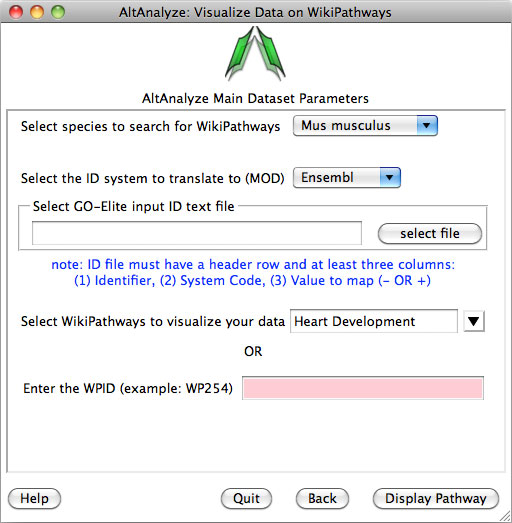
Once this menu appears:
- Select the species in which your IDs are from.
- Next, select the location of the input file described above.
- Next select the name of the WikiPathway of interest to color. If you just created the WikiPathway at http://wikipathways.org/index.php/WikiPathways, you may need to wait a few hours for it to appear in the pull-down menu.
- You can also optionally enter the WikiPathway ID directly.
- After selecting "Display Pathway", you will wait for up-to 30 seconds for the pathway to appear. A PDF and PNG version will be saved to the folder "WikiPathways" in the location where the input file was selected.
Pathway Analysis Menu
All enriched WikiPathways can also be automatically generated following the GO-Elite biological enrichment analysis in AltAnalyze. To do this, simply select the option Visualize all over-represented WikiPathways from the Pathway Analysis menu under Additional Analyses after selecting the folder containing your input text file(s). Each colored pathway will appear in the folder WikiPathways in the input folder selected.
Pathway Visualization Outside of AltAnalyze
More advanced options for pathway visualization can be achieved in a number of external analysis tools. This will require additional import options, however, AltAnalyze already generates an input file for these applications in the folder ExpressionOutput, with the file prefix GenMAPP. We recommend the applications PathVisio and GenMAPP-CS for this purpose.
PathVisio
PathVisio, like GenMAPP-CS, can be used to load pathways from WikiPathways. In addition, it can create new pathways and has a number of sophisticated options for streamlined data visualization.
The same input file used for GenMAPP-CS can also be loaded into PathVisio. This will require that the text file is imported and processed before creating criterion. For example directions, see their tutorial here.
GenMAPP-CS
GenMAPP-CS can be downloaded from http://www.genmapp.org/beta/genmappcs/. Compatibility and installation information can be found here.
To load the GenMAPP input text file produced by AltAnalyze (ExpressionOutput directory), see the following tutorials.