Once alternative probe sets have been identified from an AltAnalyze exon array analysis, you can easily load this data in the Cytoscape plugin DomainGraph to:
-
View prioritized AltAnalyze highlighted alternative exons and annotations.
-
Assess which probe sets overlap with a set of loaded genes and which specific protein domains at a high-level (protein/domain/miRNA binding site network/pathway) and low-level (domain/exon/probe set view).
-
View alternative probe set data in the context of WikiPathways and Reactome gene networks.
Figure 8.1
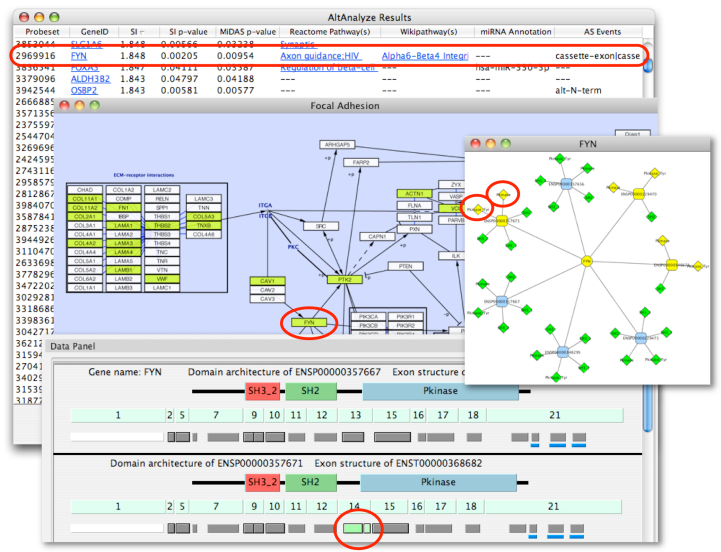
Visualization of AltAnalyze regulated probe sets along exons and protein domains: In the top panel, a loaded Cytoscape DomainGraph network is shown for the gene FYN, with relevant protein domain interactions shown between two alternative isoforms of each gene. Rounded boxes represent gene nodes and diamonds, protein domains and other functional elements. Greenish yellow nodes represent those containing AltAnalyze regulated probe sets, whereas green do not overlap with an AltAnalyze regulated probe set. The gene FYN has been selected in the main network that creates a domain architecture and exon structure view for the select FYN isoform in the Cytoscape "Data Panel". Domains (top), exons (middle) and probe sets (bottom) are shown that correspond to the FYN isoforms ENSP00000229470 and ENSP00000229471, with AltAnalyze down-regulated probe sets in green. Probe sets with a solid black border are associated with an alternative splicing (alternative cassette exon) or alternative promoter annotation. In this example, the probe set with an alternatively splicing annotation overlaps with exon 8 and the Protein Kinase domain of the protein. Probe sets with a blue bar beneath them overlap with predicted microRNA binding sites. Details about each domain, exon, probe set and microRNA binding site, including AltAnalyze statistics and functional annotations are accessible by mousing-over the respective feature and by left-clicking the object to link-out to resources on the web.
Installing DomainGraph with AltAnalyze
-
Simply download and extract AltAnalyze 1.15 (or higher) from the compressed installation file and Cytoscape, DomainGraph and the GPML puglin will be ready to use. This plugin can be updated through the Cytoscape plugin manager.
-
Start Cytoscape directly from AltAnalyze after the alternative exon results are produced. Alternatively, open Cytoscape manually (AltAnalyze_v1release/Cytoscape).
-
Download the species gene database of interest (required for the first use - see below).
Running DomainGraph
Detailed instructions on running DomainGraph can be found here or in the AltAnalyze application folder: AltAnalyze_release/Documentation/domain_graph.pdf.
Platform Compatibility
The DomainGraph database is designed to specifically work with Affymetrix Exon 1.0 arrays, however, AltAnalyze exports files for the Affymetrix Gene 1.0, HJAY and MJAY arrays as well as human, mouse and rat RNA-Seq results that allow for visualization of these results as well. In order to accomplish this, non-exon array probe set IDs are converted to exon array probe set IDs. This translation is performed by matching the optimally aligning exons, junctions or probe sets using the AltAnalyze exon block/region annotations and probe set genomic positions. For reciprocal probeset junction array analyses (ASPIRE or Linear Regression), matching is to the predicted alternative exon rather than the reciprocal junctions.