Marker Discovery Analysis in AltAnalyze
AltAnalyze includes a built-in method to identify putative markers that are selectively expressed in a single analyzed condition. This method is most appropriate when analyzing greater than 2 biological groups to discover genes or other IDs that can optimally distinguish a given biological group from all others. MarkerFinder is run by default in the standard AltAnalyze workflows.
This approach is distinct from the LineageProfilerIterate approach, which allows for more sophisticated marker identification, along with training and evaluation methods.
Core Algorithm
The core algorithm is the same one used for the LineageProfiler cell-type marker discovery pipeline as described here.
Default Analysis
By default, MarkerFinder is run when performing a standard AltAnalyze expression Analysis. Both tabular and heatmap image files will be produced. The tabular files are saved to the folder ExpressionOutput/MarkerFinder and the heatmap image files are saved to DataPlots/MarkerFinder.
Application of MarkerFinder Results for Sample Classification
MarkerFinder result files can be loaded as input in the LineageProfiler analysis menu. First, download AltAnalyze from http://www.altanalyze.org, extract to your hard drive and install the latest human database when prompted (currently EnsMart65) after running the first time. From the Main Menu select “vendor/data type” as “Other ID” and under “platform” select “Symbol”, then select “Continue”. Next, select “Additional Analyses” then select “Continue”. Next, select the menu item “Lineage Analysis” and “Continue”.
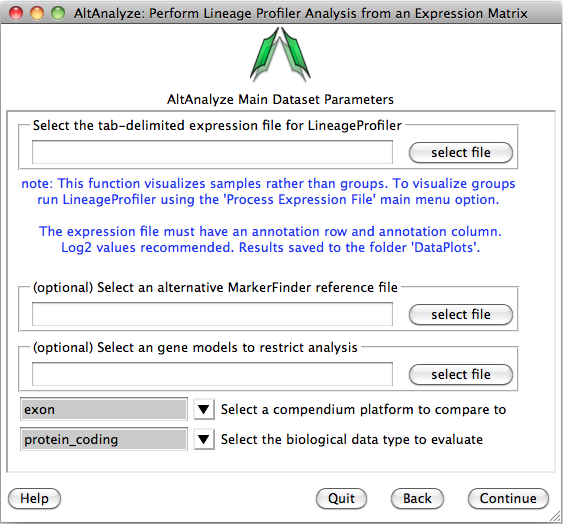
You will be presented with the analysis window to upload you expression (e.g., delta CT) values for your analyzed set of IDs (e.g., genes) and a reference expression file containing your two classes (e.g., cancer and wt) (“Select an alternative MarkerFinder reference file”). Next, you can optionally select the option “Select marker models to restrict analysis to”, to select a file with different ID sets to analyze. Leave the other options as default and select continue to run.
Additional details can be found here.